Bacteria are much more complex than we give them credit for say scientists studying how pathogenic bacteria use a process known as “cleavage” to expand the functional capacity of proteins. They hope their insights into how and where cleavage occurs will shed light on the critical functional motifs within proteins and gain deeper insight into the functional complexity of proteins. These studies will underpin efforts to hasten develop of vaccines urgently needed to protect both human and animal health.
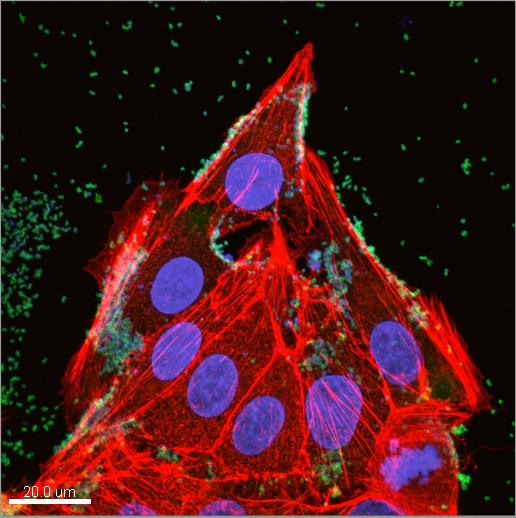
Image Credit: Ben Raymond/ Caption: Characteristic depiction of Mycoplasma hyopneumoniae cells (green circles) adhering to porcine epithelial cells (stained in red and blue). M. pneumoniae is the second most frequent cause of bacterial pneumonia and the emergence of antibiotic-resistant strains is a worldwide problem in clinical medicine.
The results of international collaborations, published in two Scientific Reports papers, is giving a better understanding of how pathogenic bacteria such as Mycoplasma hyopneumoniae, Mycoplasma pneumoniae and Staphylococcus aureus use enzymes to cleave proteins into smaller functional pieces. Protein processing represents a novel mechanism to unlock the full functional capabilities of proteins that traffic to different cellular locations. The researchers say that cleavage can re-define and expand the functional capacity of proteins.
Led by University of Technology Sydney (UTS) ithree institute scientists, new chemical tagging methods helped identify the extent of protein cleavage in M. hyopneumoniae, a bacteria causing huge losses in the swine industry worldwide. The research team was surprised to find that many proteins, particularly those on the bacterial cell surface were a target of processing events. These cleavage fragments are then able interact with the host.
“Bacteria continue to develop resistance to many commonly used antimicrobials, so we desperately need to learn more about these bugs in order to create new therapeutics,” says Iain Berry, a PhD candidate at UTS ithree institute and co-lead author on one of the papers.
“Our research into M. hyopneumoniae means that the proteins actually responsible for the cleavage can now be targeted and the cleavage process blocked.”
Authors Michael Widjaja, Kate Harvey and Lisa Hagemann say their research shows that ancient proteins in these pathogens have evolved to be multifunctional.
For the first time the research was also able to show how a protein called “elongation factor Tu”, which is found in all bacteria, can display multiple functions. The idea that this moonlighting protein is a target of processing events that generate fragments that retain moonlighting functions, raises new questions about the role of protein cleavage and how it might serve to expand protein function. M. pneumoniae is the second most frequent cause of bacterial pneumonia and the emergence of antibiotic-resistant strains is a worldwide problem in clinical medicine. S. aureus is also a major pathogen that has developed the ability to resistant antibiotics. We are yet to understand the implications of protein processing in the pathogenesis of these important pathogens.
“Processing has implications for how these pathogens establish infection and it’s a concept that may now be investigated in a broader selection of pathogens,” they say.
The research teams believe that the methodologies developed can also be applied to other bacteria.
The research was funded through Ausgem , UTS and the Deutsche Forschungsgemeinschaft. Ausgem is a collaborative partnership between the NSW DPI Elizabeth Macarthur Agricultural Institute (EMAI) and the UTS ithree institute.
Publication details:
Paper 1: N-terminomics identifies widespread endoproteolysis and novel methionine excision in a genome-reduced bacterial pathogen, Iain J Berry et al, Scientific Reports 2017 doi: 10.1038/s41598-017-11296-9
Paper 2: Elongation factor Tu is a multifunctional and processed moonlighting protein Michael Widjaja, Kate Harvey and Lisa Hagemann Scientific Reports 2017 doi:10.1038/s41598-017-10644-z

